Thoughts on Julia and R
Hello, Julia
Julia is an exciting new technical computing language. It’s still in its infancy, but it’s fast (see below), and already does a lot.
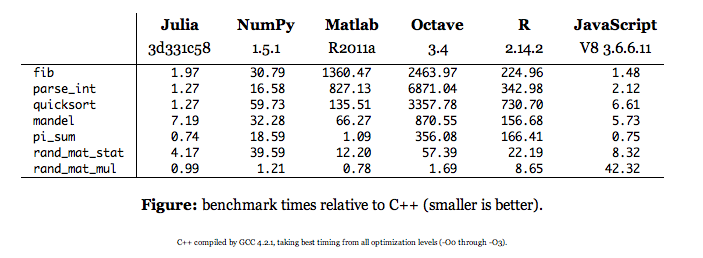
There’s been some excitement on Twitter about Julia. Excitement combined with open source often yields development, which then leads to further excitement, until a mature open source project arises. One of Julia’s explicit goals is to challenge other statistical computing environments, including R.
What’s wrong with R?
R is, without a doubt, changing the world. It’s being used by industry giants like Facebook and Google, while also providing academic researchers in statistics, biology, psychology, and countless other fields with not only a free and open source statistical environment, but a huge number of user-contributed package through CRAN. Now methods papers in many fields are often accompanied by CRAN or Bioconductor packages. It’s also a brilliant platform for reproducible, open research, as Bioconductor beautifully illustrates with packaged and version-controlled genomes, microarray probesets, etc.
However, R is suffering from growing pains. For example, there are now 64-bit versions of R, however, vector indexing is still limited by R_len_t (see definition in src/include/Rinternals.h):
/* type for length of vectors etc */
typedef int R_len_t; /* will be long later, LONG64 or ssize_t on Win64 */
#define R_LEN_T_MAX INT_MAXIt appears that one can simply change this to a long and recompile to increase the longest possible addressable vector, but no. Take a look at R_euclidean in library/stats/src/distance.c for an example why: almost all variables for iterating over elements in vectors are defined as integers and don’t use this type. One would have to read through every function, and every line of code to fix this.
R_len_t is just one example. Another issue is that R has been slow to adopt new compiler technologies (i.e. JIT, optional type indications, etc). R almost always gains speed from pushing stuff to C (the recent bytecode compiler is an exception). This isn’t a problem, but it’s a huge limitation to require developers to not only know R, but also C, and also how to interface the two. More modern languages (Java, as well as Python and Julia come to mind) spend more time tracking compiler technology developments and implementing them than R core does (again, Luke Tierney and the bytecode compiler are exceptions). It’s still sometimes efficient to use C with these languages (consider Cython), but developers in these language aren’t cracking open Kernighan and Ritchie everytime they need to have a for loop do something quickly.
Another gripe I have is that R language development is somewhat closed. Despite a quickly expanding user base, the number of R core contributors is not increasing. I find it hard to believe this is due to lack of interest. It seems much more likely this is due to institutional reasons that need to be changed. The nice thing about language development that it’s really hard, so opening up R to more contributors won’t likely flood the existing core with bad ideas and patches. Personally I would dedicate much more time profiling, reading the source, and working on the R language if it were more open.
The last gripe I have is that R is fragmented. Consider Python:
import re
re.search(r'R-([\d]+).([\d]+)', "R-2.15").groups()Now, consider R:
gsub("R-([\\d]+)\\.([\\d]+)", "\\1", "R-2.15")
# or
library(stringr)
str_match("R-2.15", "R-([0-9]+)\\.([0-9]+)")Now, Python also has PyPI’s re2, but most developers are using re. The motivation behind stringr is that R’s currently family of string processing functions are horribly inconsistent:
# (my ... to avoid writing all parameters)
grep(pattern, x, ...)
regexpr(pattern, text, ...)
gsub(pattern, replacement, x, ...)
strsplit(x, split, ...)But rather than deprecate these and move forward, we now have two sets of string processing functions. Both are being used. I’m not saying Hadley Wickham is to blame here; quite the contrary, he’s trying to fix a very annoying problem in the language and should be commended. I think the community needs to be more open; for example, before writing a package that processes strings, let’s discuss an implementation plan, deprecating old functions, etc. If not, in the future R will be highly fragmented, and end up with five different object orientation systems… oh, wait.
What would it take to “challenge” R?
Contributors to Julia are optimistic they can challenge R based on a solid foundation of JIT compiling, parallelism, and nice language semantics. I salute this optimism, but I think we need to realistically consider what it would take to “challenge” R.
First, we would need to build an equal statistical computing environment. Consider moving all of stats, MASS, graphics, grid, etc to Julia. Is Julia sufficiently faster than R will be in the time it takes to port these base packages? Remember, R is a moving target; despite my few earlier gripes, R will evolve and get faster. Now, consider adding the extremely popular CRAN packages like ggplot2 and lattice to Julia. In the time it takes to port these, is Julia still sufficiently faster than R will be?
Suppose it is still faster than R. What about after we port the rest of CRAN, and all of Bioconductor to Julia? My point isn’t say that it’s unimaginable that Julia will surpass R. It’s that developers should really dissect what makes a successful language successful before they try to challenge it. I don’t have a horse in this race; I would love to see Julia surpass R. But if all developers want is a fast environment to analyze large data sets using a wealth of methods and libraries, it may be a lot easier to make R faster than to develop a new fast language and hope/wait/beg the community to move over.
Update: Julia core developer Jeff Bezanson sent me a very kind email on March 9th, 20012 about this post. In it, he said the “challenge R” statement was made by a community member and is in no way the mission of the Julia language. He had many kind words to say about the R langauge and its statistical functionality.