The Beauty of Bioconductor
In talking with bioinformaticians, biologists, and other researchers, I’ve seen some worrying trends in computation in the sciences. I plan on writing about these extensively in the future, as I believe computation in the sciences will not scale well to face the huge wealth of data coming experiments will provide. This is not due to algorithmic or hardware limitations, but rather to the fact that scientific programmers simply do not have the same standards and practices that the software industry does.
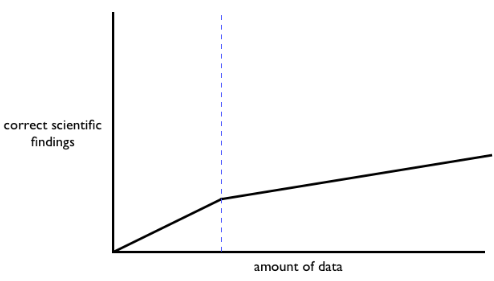
Three events are simultaneously occurring that could endanger the validity of scientific conclusions in the future. First, new technology is providing the average scientist with more data than ever before. Genomics is the prime example of this: the average biologist can now sequence multiple samples simultaneously whereas this would be prohibitively expensive just a few years earlier. As metabolomic and proteomic data are increasingly incorporated into research alongside genomic data, the work done by bioinformaticians will increase significantly. More lines of code to write and more data to process under deadlines will doubtlessly lead to mistakes.
The second contributing factor is that more researchers are writing code and analyzing their own data rather hiring a bioinformatician or statistician. It’s an awesome and commendable occurrence, but sadly academic institutions don’t adequately prepare researchers to code to high standards. Also, in many cases these researchers learn to program by analyzing their own experimental data, rather than example or “toy” data. This makes “silent” mistakes (i.e. those that don’t prompt an error, but lead to incorrect results) impossible to discover as the actual results are not known.
The last contributing factor is that there’s not a strong expectation that coding standards and software engineering practices be upheld in the sciences. There’s a strong cowboy coding culture in scientific programming. In this mindset, the coding is done when the data is processed, not when the data is processed, the code documented, the unit tests passed, the code checked into a repository, etc. The scientific community needs to embrace the idea that proper data analysis takes time: perhaps as long or longer than gathering experimental data.
In future essays I’ll talk more about these issues in depth. This stuff honestly keeps me (and other people I know) awake at night. I worry humanity may face a Thalidomide-like event in the future due to an error in scientific programming.
However, here I want to commend a project that I feel is underutilized in the biology and bioinformatics communities: Bioconductor. It’s worthy of praise as both an example of, and tool to aid in excellency in bioinformatics programming. I’ll focus primarily on its capacity for handling high throughput sequencing data (even though it handles data from other assays very well too).
Where is Bioinformatics?
Currently, many bioinformatics analyses go something like this: experimental data is received, and then a bioinformatician downloads vast amounts of other data relating to the experiment from web resource such as the UCSC Genome Browser, Ensembl, Phytozome, etc. Often this includes genome assemblies, transcript sequences, and annotation data. Then, application code (alignment software, assemblers, SNP finding tools, etc) are downloaded and compiled. These tools are used alongside custom code written that combines downloaded data with the experimental data, and this produces results that are interpreted. Intermediate results may be fed into other online tools and databases like DAVID or Reactome.
However, this is a bad model if one wants the analysis to be reproducible. The common weakness is that web resources can be unstable. It’s then necessary for the researcher to record software and data versions manually. Even if the researcher dutifully complies, outside databases and code repositories may disappear and leave the project unable to be reproduced. Researchers truly invested in conducting reproducible research then have to store data and software versions themselves, which given the scale of genomic data is quite a burden.
Thus, three things currently perplex reproducible research in bioinformatics: the scale of both experimental and other required data prevents easy self-archival, the fast-paced development of bioinformatics tools could lead to differing results across versions, and the overwhelming prevalence of web-based data resources and applications which are not easily reproducible.
The Bioconductor Solution
Bioconductor has, in my opinion, the best solution to these problems. First, Bioconductor stores past versions of its packages back to their earliest releases. Past experiments can be replicated using the exact version of software that was used for the actual analysis.
Second, Bioconductor stores data as packages. Pre-packaged versioned data is a cornerstone of reproducible research. For example, suppose I am working with human RNA-seq data. This requires transcript annotation data, which could be downloaded from an online resource. To be reproducible, this requires that:
The webhost be up indefinitely.
The URL remain stable and point to the exact same resource.
The user provide not only a URL but the version of data/software downloaded.
That the external resource provider (i.e. database or application developer) actually update their versions accordingly.
For absolute best practice, it’s also necessary to MD5 checksum the data and record this value to maintain any data gathered from the same source is the exact same.
In contrast, consider how I would load human transcript data into R with Bioconductor:
library(TxDb.Hsapiens.UCSC.hg19.knownGene)The versioning here is done explicitly in the package name: hg19. I could also easily record the state of all my Bioconductor packages and my session with sessionInfo():
> sessionInfo()
R version 2.14.1 (2011-12-22)
Platform: x86_64-apple-darwin9.8.0/x86_64 (64-bit)
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
loaded via a namespace (and not attached):
[1] annotate_1.32.2 AnnotationDbi_1.17.23 Biobase_2.14.0
[4] BiocGenerics_0.1.12 DBI_0.2-5 DESeq_1.6.1
[7] genefilter_1.36.0 geneplotter_1.32.1 grid_2.14.1
[10] IRanges_1.12.6 RColorBrewer_1.0-5 RSQLite_0.11.1
[13] splines_2.14.1 survival_2.36-12 xtable_1.7-0 Entire genomes are also packaged via the BSgenome package (BS refers here to Biostrings). If the data in packages is not sufficiently recent, the GenomicFeatures package provides a programmatic way of downloading, packaging, and using data from BioMart and UCSC Genome Browser tracks, and provides functions for saving and loading transcriptDb objects from such resources. Recently Duncan Temple Lang and I were speaking about reproducible research, and he said “people adopt best practices when they’re right in front of their face”. Bioconductor’s tools do just that. Furthermore, Bioconductor has strict coding and documentation standards (much stricter than CRAN actually), which ensures user-contributed packages are high quality.
Information leakage and statistics at every level
When discussing R and Bioconductor with other researchers, it’s easy to convince them to adopt both for analyzing statistical data - the data that comes in the very final stages of a bioinformatics analysis. It’s usually much more difficult to convince them to consider working with high throughput sequencing data in Bioconductor. Folks complain that it’s (1) not worth it to process sequencing data with Bioconductor tools or (2) it’s not fast enough. I’ll address the second point in a bit; more importantly I want to emphasize that it’s absolutely worth it to process sequencing data in Bioconductor.
In analyzing genomic data, we take very, very, very high dimension data and try to condense it into biologically meaningful conclusions without being misleading or getting something wrong. Every step is about taking dense data and making it understandable: we take sequence reads and try to assemble them into larger contigs and scaffolds, we take cDNA reads and try to map them back to genomes to understand expression, etc. At each step, our tools make heuristic or statistical choices for us. Pipelines woefully ignore these choices because in most cases, after a step is completed, a script jumps to the next step.
When I think about these steps, I try to assess what I think of as “informational leakage” in bioinformatics processing. Each step summarizes something, hopefully in a way without bias or too much noise. Informational leakage is the information that’s lost between steps. Catastrophic information leakage occurs when we lose information that could have indicated whether the data is biased or incorrect. We can hedge the risk of information leakage when we use summary statistics between steps that try to capture this leaked information.
Consider processing RNA-seq reads. The first step is usually quality control, i.e. removing adapter sequences and trimming off poor quality bases. Failing to gather summary statistics before and after each of these steps leads to potentially catastrophic information leakage. Suppose that an experiment with control and treatment groups, sequenced on two different lanes (bad experiment design!). If one lane has systematically lower 3’-end quality than the other, quality trimming software will trim these bases off and lead one experimental group to have much shorter sequences than the other. The mapping rates will differ significantly, as shorter reads may map less uniquely. In the end, our data is completely confounded not only by the lane (and bad experimental design), but by our own tools! If these tools are being run in a pipeline without intermediate summary statistics being gathered, this catastrophic information leakage will go unnoticed.
Loading sequencing data into R and using Bioconductor’s tools earlier allows summary statistics to be gathered earlier and more easily (R is, after all, great for statistics and visualization), which I strongly believe will decrease the risk of catastrophic information leakage in genomics data analysis. This is why I wrote qrqc, which can provide quick summary statistics on sequencing read quality. Used before and after the application of quality tools, qrqc can provide information not only on the state of the data, but also the effect of the tools.
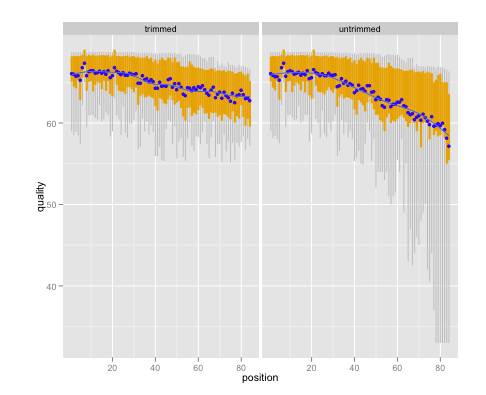
With existing Bioconductor packages, many useful statistics can be gathered on whole reads, BAM mapping results, VCF files, etc.
Massive Power
The complaint that R is slow, and couldn’t possibly be used with sequencing/mapping-level data is unwarranted. In reality, some of Bioconductor’s core packages for working with high throughput sequencing data such as Biostrings and IRanges (the foundation of GenomicRanges) are astoundingly fast because most of their backends are written in C. Biostring actually uses external pointers to C structures and bit patterns to encode biological string data efficiently.
In addition to being fast, they’re also clever. Biostrings implements an abstraction called a “view” on an XString object, which efficiently represents multiple sections of the same string object (such as subsequences of interest). While I wouldn’t write a short read aligner or assembler entirely in R, many bioinformatic tasks are more than sufficiently fast with Bioconductor tools.
Conclusion
In evangelizing Bioconductor, I have two goals. First, I want to spread awareness that it’s the best way to do reproducible bioinformatics that I think exists today. I want more people to use it just because I deeply care about the state of science and reproducibility. Second, I want to build excitement about this project so that more people will contribute. I believe that far too many high quality bioinformatics tools are written outside of Bioconductor. Packaging bioinformatics tools in Bioconductor forces the developer to adopt strict standards, write clear documentation, and open up a program to a large, active user base. Any results from a package’s methods can then easily be evaluated using R, CRAN, or Bioconductor’s existing tools.
I also believe that large programs (like BLAST, and maybe assemblers) should provide better interfaces to R, to prevent information leakage in analysis. I’m willing to bet that a large majority of bioinformatics tools could be outputting more statistics than they currently do that could be valuable in assessing their functionality. R interfaces to these bioinformatics tools will drastically make it easier for biologists and bioinformaticians to prevent information leakage.